# Reorder factors
gene_exp$Dose <- ordered(gene_exp$Dose, levels=c("Ctrl", "Gy10"))
gene_exp$Time <- ordered(gene_exp$Time, levels=c("2h", "6h", "24h"))
gene_exp_summary$Dose <- ordered(gene_exp_summary$Dose, levels=c("Ctrl", "Gy10"))
gene_exp_summary$Time <- ordered(gene_exp_summary$Time, levels=c("2h", "6h", "24h"))
# Line plot ----
pd <- position_dodge(0) # avoid errorbar overlap
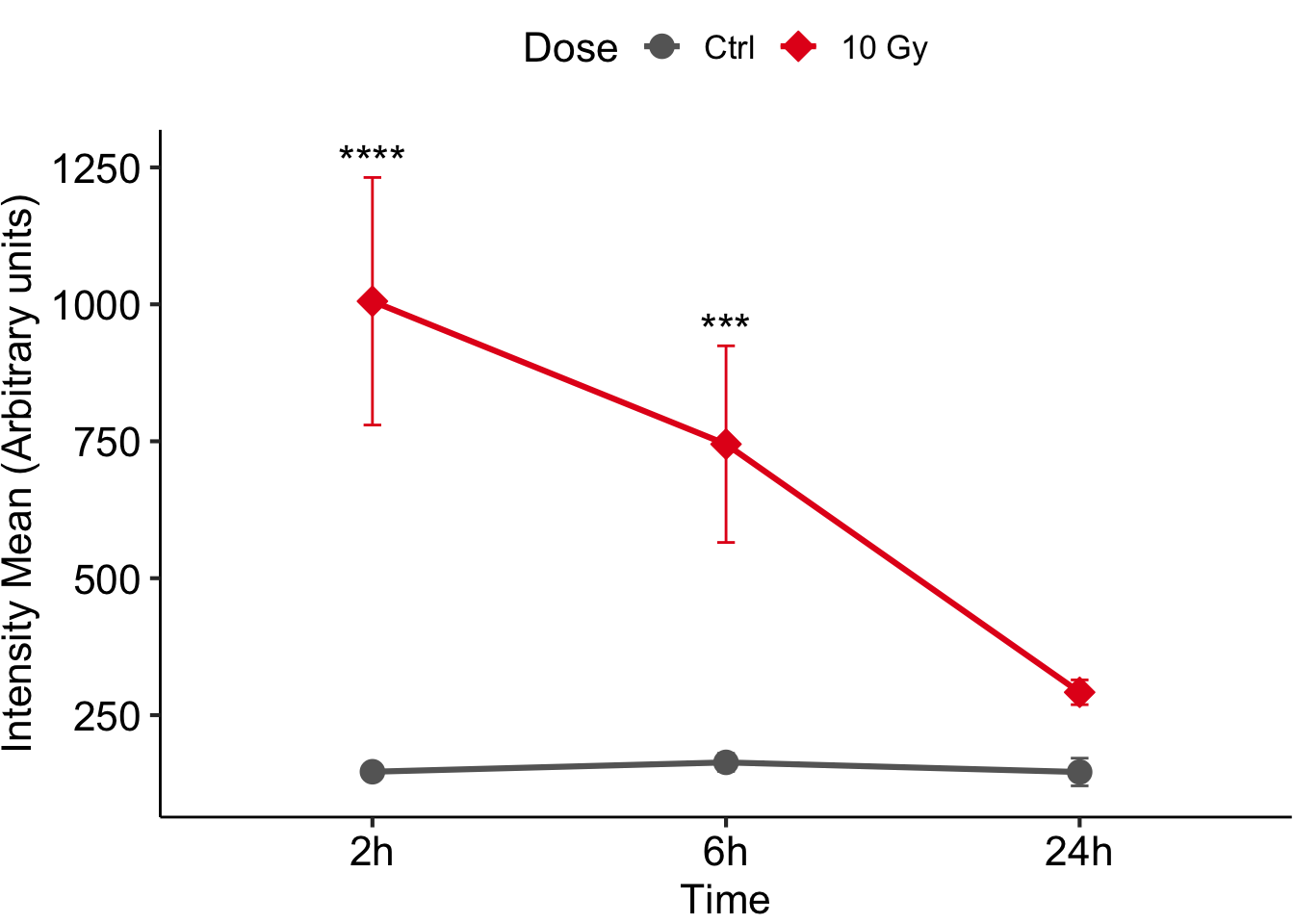
figure <-
ggplot(data=gene_exp_summary, aes(x=Time, y=mean, colour=Dose, group=Dose)) +
geom_line(size=1.1, position=pd) +
geom_point(size=4, aes(shape=Dose, fill=Dose), position = pd) + #oppure +geom_point(size=5, shape=21, fill='white')
geom_errorbar(aes(ymin=mean-sd, ymax=mean+sd, colour=Dose), width=0.1, position = pd) +
ylab("Intensity Mean (Arbitrary units)") +
scale_shape_manual(values=c(21,23), labels = c("Ctrl", "10 Gy")) +
scale_color_manual(values=c("grey40", "#E41A1C"), labels = c("Ctrl", "10 Gy")) +
scale_fill_manual(values=c("grey40", "#E41A1C"), labels = c("Ctrl", "10 Gy")) +
annotate("text", x = c(1,2), y = c(gene_exp_summary$ypos[5], gene_exp_summary$ypos[6]),
label = c("****", "***"),
size = 6) +
theme_pubr(base_size = 16, margin = FALSE, legend = "top")